Using NanoString Data to Identify Immune Signatures in Patients With Mild to Moderate, Severe or Critical Cases of COVID-19
The following case study features published data from Hadjadj et al., 2020, Science.
Study Aim
This study aimed to identify characteristic immune signatures in whole blood samples from patients at Cochin Hospital (Paris, France) with “mild to moderate”, “severe” or “critical” cases of COVID-19. Samples were taken eight to twelve days after the onset of symptoms. Expression of 579 immunology-related genes was evaluated using the NanoString Human Immunology Panel v2, as described in Hadjadj et al., 2020, Science. The dataset was provided to Fios Genomics by the authors of this publication. Gene expression and immune signature scores were evaluated for each of the severity groups, and compared to a set of healthy controls.
Analysis and Results
Fios bioinformaticians independently reanalysed the NanoString dataset to validate and expand upon the conclusions of the original study. Our team performed association analysis for differentially expressed genes and gene signatures, along with functional enrichment analysis to identify pathways and GO terms that were associated with disease severity.
These data analyses revealed that more serious COVID-19 cases typically have lower activity of the adaptive immune system and possibly increased activity in the innate immune system.
Analysis of gene expression data also revealed widespread changes in the levels of immune markers such as TLR5, BST1, LILRA5 and MAPK14 in COVID-19 patients compared to healthy controls.
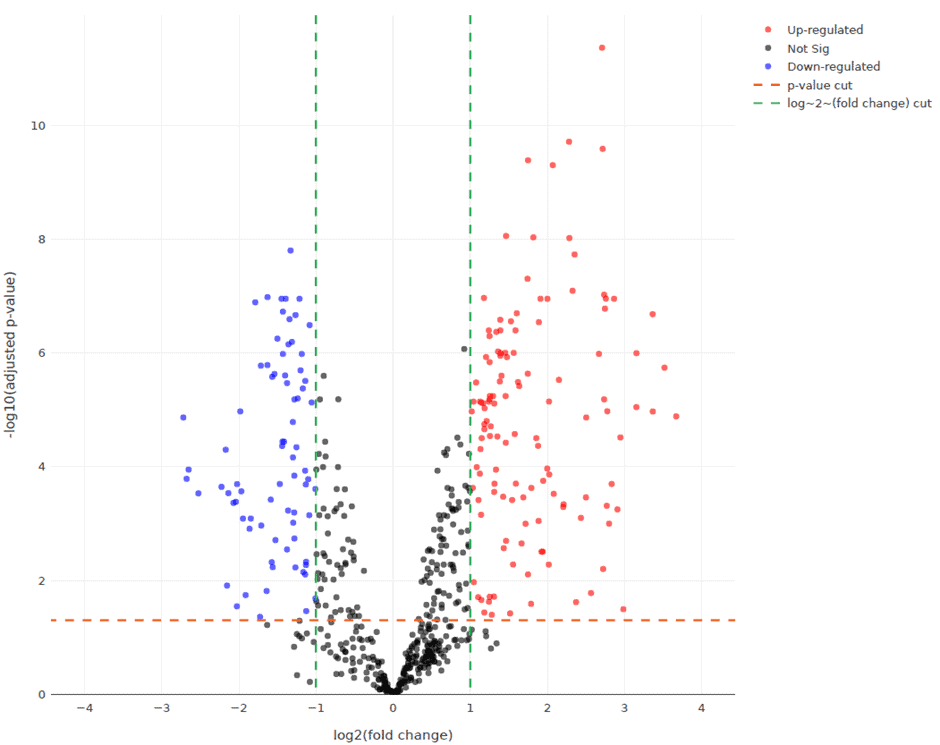
TLR5 recognises bacterial flagellin and may reflect the presence of secondary bacterial infections in patients with severe and critical cases of COVID-19. Immune cell gene signatures showed significant reductions in the number of T cells, B cells, natural killer cells and cytotoxic lymphocytes in critical cases of COVID19.
Changes were also observed in the activity of key immune pathways including interferon, interleukin, T-cell receptor (TCR), toll-like receptor (TLR) and major histocompatibility complex (MHC) signalling, as shown in the figures below:
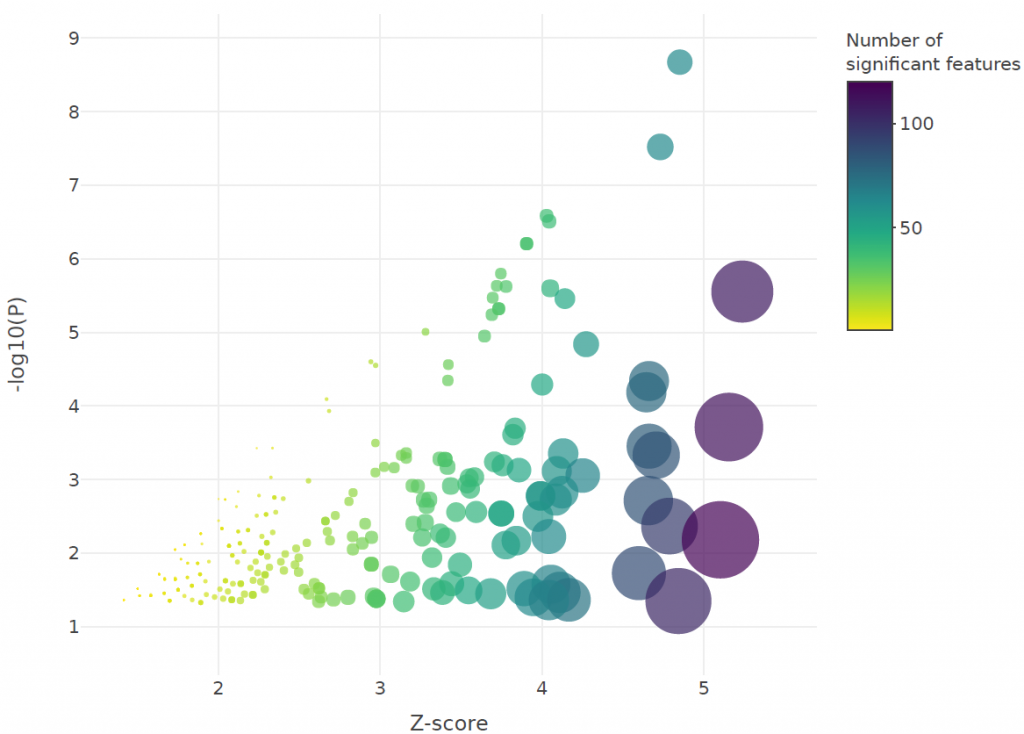
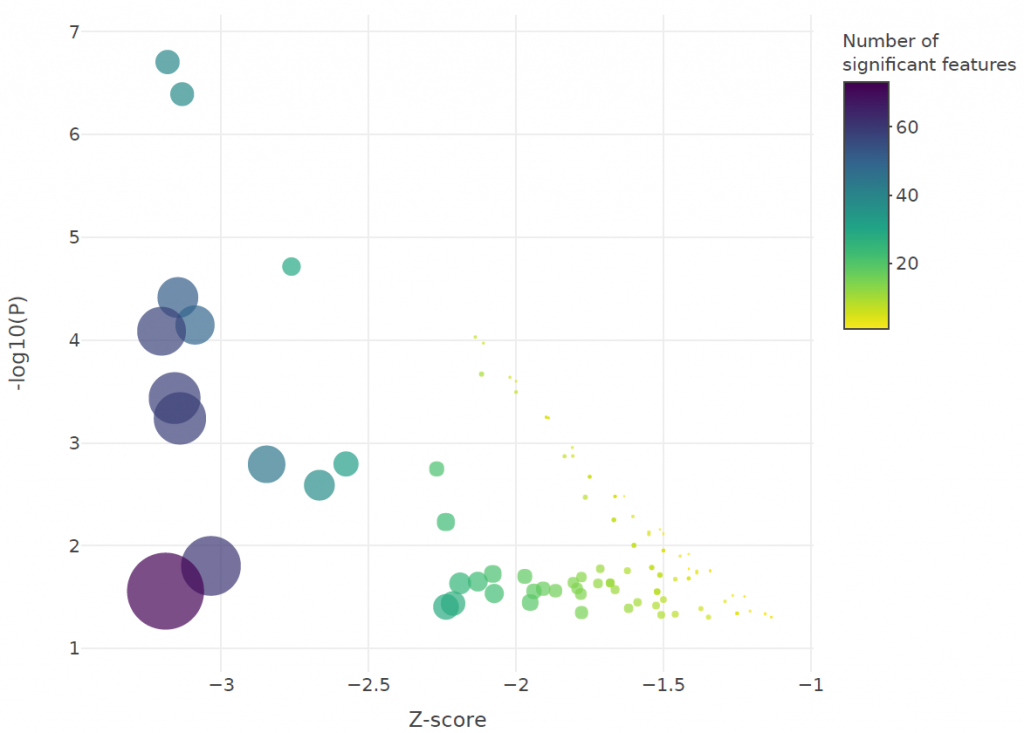
Conclusions
The team found that pathways involved in the adaptive immune response as well as interleukin and interferon signalling, were more likely to be enriched in “mild to moderate” cases, whereas early immune pathways such as TLR signalling, were enriched in severe and critical cases. We also found evidence that more serious COVID-19 cases are associated with a tendency towards innate rather than adaptive immunity. We recommend follow-up studies to validate the attenuation of adaptive immunity in COVID-19 patients and to unravel the mechanism behind it.
View more case studies and publications.
Request a Sample of our NanoString Data Analysis Reports
Fill in the form below to access the data analysis report our team created. Here, bioinformatics analysis helped to identify characteristic immune signatures in whole blood samples. Expression of 579 immunology-related genes was evaluated using NanoString datasets.

