Pharmacological Inhibition of PMS2 Increases Tumor Mutational Burden, Induces Microsatellite Instability and Elicits Immune Mediated Rejection in vivo
The following case study features data from our collaboration with NeoPhore which was presented at the EORTC 2023 (1) and AACR Annual Meeting 2024 (2).
Background
DNA mismatches arise during cell replication through the incorporation of an incorrect base in the nascent DNA strand or a slippage of the replication machinery on repetitive DNA, such as microsatellites, that subsequently escape DNA polymerase proofreading. These replication errors are usually recognised and corrected by DNA mismatch repair (MMR) proteins but go unrepaired when MMR is impaired. MMR deficiency (MMRd) therefore leads to elevated mutational burden and expression of neoantigens on the cell surface of dividing cells over time. MMRd tumours, observed at varying frequency in different cancer types, have been shown to exhibit sensitivity to immune checkpoint inhibitor treatment compared to MMR proficient tumours and mutation-associated neoantigen expression has been correlated with improved immune surveillance and progression free survival (3, 4, 5). The knowledge of a tumour’s MMR status may therefore be informative for clinical treatment decisions. As such, approaches are sought to induce MMRd thereby rendering a tumour responsive to immunotherapy. NeoPhore is developing MMR modulators, that pharmacologically inhibit MMR proteins and induce checkpoint inhibitor sensitivity (6).
Study Aim
The aim of this study was to investigate if treatment with NP1867, NeoPhore’s proprietary small molecule PMS2 MMR protein inhibitor, increased tumour mutational burden (TMB) and microsatellite instability (MSI) over time.
Experimental Design, Data Analysis and Results
Experiments were performed by NeoPhore by treating the murine colorectal cancer cell line CT26 with NP1867. Three replicates per treatment duration were sequenced using whole exome sequencing.
Fios Genomics performed sequence read alignment, variant calling according to GATK best practices and determined the proportion of microsatellite instable (MSI) homopolymer and microsatellite sites (7) per sample. We then investigated the time-dependent accumulation of mutations such as single nucleotide variants, insertions and deletions, the overall TMB, the TMB gain from baseline (untreated), and the MSI status for each sample. We observed an increased TMB from baseline (Figure 1a) and elevated MSI (Figure 1b) with NP1867- compared to DMSO-treatment over time, indicative of MMRd.
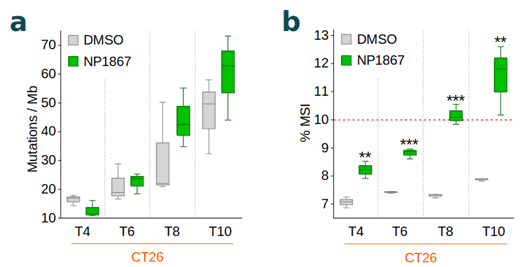
Conclusions
Our analysis provides evidence for the emergence of mutational events consistent with MMR deficiency. These results correlated with an increased sensitivity of NP1867-treated CT26 cell lines to anti-PD-1 checkpoint inhibitor treatment over time.
References
- J. Blagg, P. Riou, A. Hervieu, E. Piumatti, G. Rospo, S. Arunachalam, B. Meier, S. Weeks, M. Rodriguez, K. Parmar, P. Patel, A. Peall, P. Tongue, T. McLaren, R. Workman, R. Bago, S. Robjohns, R. Beaumont, M. Briggs, G. Langley, C. Nichols, D. Clark, P. Winship, B. Rousseau, M. Baker, M. Drysdale, G. Germano, A. Bardelli. NP1867, a potent, selective, covalent small molecule inhibitor of DNA Mismatch Repair (MMR) protein PMS2, functionally inhibits MMR in cells and elicits COSMIC mutational signatures consistent with MMR-deficient patient samples [abstract]. In: Proceedings of the AACR-NCI-EORTC Virtual International Conference on Molecular Targets and Cancer Therapeutics; 2023 Oct 11-15; Boston, MA. Philadelphia (PA): AACR; Mol Cancer Ther 2023;22(12 Suppl): Abstract nr C086. https://aacrjournals.org/mct/article/22/12_Supplement/C086/730739/Abstract-C086-NP1867-a-potent-selective-covalent
- E. Piumatti, A. Hervieu, P. Riou, J. Blagg, A. Peall, S. Weeks, M. T. Rodríguez-Plata, G. Rospo, S. Arunachalam, B. Meier, P. Tongue, T. McLaren, K. Parmar, P. Patel, D. Clark, G. Langley, C. Nichols, B. Rousseau, P. Winship, M. Baker, M. Drysdale, G. Germano, A. Bardelli. Pharmacological inhibition of PMS2 increases tumor mutational burden, induces microsatellite instability and elicits immune mediated rejection in vivo [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2024; Part 1 (Regular Abstracts); 2024 Apr 5-10; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2024;84(6_Suppl): Abstract nr 6011. https://aacrjournals.org/cancerres/article/84/6_Supplement/6011/735902/Abstract-6011-Pharmacological-inhibition-of-PMS2
- D. T. Le, J. N. Uram, H. Wang, B. R. Bartlett, H. Kemberling, A.D. Eyring, A.D. Skora, B.S. Luber, N. S. Azad, D. Laheru, B. Biedrzycki, R.C. Donehower, A. Zaheer, G. A. Fisher, T. S. Crocenzi, J. J. Lee, S. M. Duffy, R. M. Goldberg, A. de la Chapelle, M. Koshiji, F. Bhaijee, T. Huebner, R. H. Hruban, L. D. Wood, N. Cuka, D. M. Pardoll, N. Papadopoulos, K. W. Kinzler, S. Zhou, T. C. Cornish, J. M. Taube, R. A. Anders, J. R. Eshleman, B. Vogelstein, L. A. Diaz Jr., PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med., 372, 2509-2520 (2015).
- V. Lee and D. T. Le, Efficacy of PD-1 blockage in tumors with MMR deficiency, Immunotherapy, 8, 1 – 3 (2016).
- G. Germano, S. Lamba, G. Rospo, L. Barault, A. Magrì, F. Maione, M. Russo, G. Crisafulli, A. Bartolini, G. Lerda, G. Siravegna, B. Mussolin, R. Frapolli, M. Montone, F. Morano, F. de Braud, N. Amirouchene-Angelozzi, S. Marsoni, M. D’Incalci, A. Orlandi, E. Giraudo, A. Sartore-Bianchi, S. Siena, F. Pietrantonio, F. Di Nicolantonio, A. Bardelli. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature, 552, 116-120 (2017).
- V. Amodio, S. Lamba, R. Chilà, C.M. Cattaneo, B. Mussolin, G. Corti, G. Rospo, E. Berrino, C. Tripodo, F. Pisati, A. Bartolini, M.C. Aquilano, S. Marsoni, G. Mauri, C. Marchiò, S. Abrignani, F. Di Nicolantonio, G. Germano, A. Bardell, Genetic and pharmacological modulation of DNA mismatch repair heterogeneous tumors promotes immune surveillance, Cancer Cell, 41, 196-209 (2023).
- B. Niu, K. Ye, Q. Zhang, C. Lu, M. Xie, M. D. McLellan, M. C. Wendl, L. Ding, MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics, 30, 1015-1016 (2014).

