RNA-Seq or Microarray: Which One Fits Best?
- 8th January 2019
- Posted by: Claudine Gabriele
- Categories: Articles, Bioinformatics, Gene Expression Analysis
Gene expression analysis
When helping with a gene expression project, one of the most asked questions is: “Which sequencing technology would be the best for my study?”
The answer is… It depends on the aim of your study and on the availability of resources. When looking at different sequencing technologies, both RNA-Seq and microarrays can be used in a gene expression study to answer a biological question. Both are connected with different benefits and challenges that this article will highlight.
Microarray technology
Microarrays can be a cost-effective choice and allow a robust outcome as microarrays involve very well-established protocols. This technology enables affordable and reliable data acquisition and still represents a good choice for genotyping-related purposes. Additionally, it is a sensible solution for when the researcher does not know which genes will be analysed.
Roughly a decade ago, microarrays used to be the standard method of choice for gene expression analysis. While they remain a key player in high-throughput technology, on the other hand, microarrays have a number of downsides; using them can restrict the researcher to detect only transcripts that correspond to what is already known in terms of genomic sequencing information. Other disadvantages are the assay’s low dynamic range and the additional requirement for probes.
When it comes to microarrays, experimental design is a key stage in order to avoid technical bias: rigorous quality control (QC) is important to identify possible hybridisation issues or platform related problems. QC is regularly performed at Fios Genomics when dealing with microarray analysis projects and other types of analysis.
What is RNA-Seq For?
The powerful features of RNA-Seq, such as high resolution and broad dynamic range, have boosted an unprecedented advance in transcriptomics research, producing an impressive amount of data worldwide.
RNA-Seq can be used on its own for transcriptome profiling or combined with other functional genomics approaches if the aim is to boost the analysis of gene expression. It can be paired with other types of assay for other purposes, such as investigating RNA–protein binding or the structure of RNA structure.
As outlined above, quality control checks should be applied at each stage of data processing before the bioinformatics analysis to ensure reproducibility and therefore increase the accuracy of the results.
There are many methods for performing an RNA-Seq experiment. In fact, the techniques are evolving so rapidly it can be difficult to decide which one to use. A very basic choice is between 1) random-primed cDNA synthesis from double-stranded cDNA, which is the most used, or 2) RNA-ligation methods (reviewed and compared in Levin 2010).
In summary, RNA-Seq is still an evolving tool but is preferable in most instances to microarrays. It is more sensitive, more robust and can be more cost-effective depending on the research’s goal.
Differential expression: With RNA-Seq you can interrogate more than just differential gene expression. Although there are microarrays available for exon-level and microRNA analysis, most users are still interested in basic, probably 3’ biased, differential gene expression. With RNA-seq you can look at coding and non-coding RNA, at splicing and allele specific expression, and possibly soon at full-length cDNA sequences, eliminating the need to infer or assemble isoforms.
Reproducibility: RNA-Seq outruns microarrays with the high degree of reproducibility it allows. This means that it is possible to reanalyse RNA-seq datasets if more information about the transcriptome is obtained.
Biases: Additionally, RNA-seq does not make use of probes or primers as microarrays do, therefore the data suffers from much lower biases. In saying this, it does not mean that RNA-Seq has no biases at all.
Read-counts: RNA-Seq provides aligned read-counts, resulting in a very wide dynamic range, improving the possibility for detection of rare transcripts.
However, in outlining the benefits of RNA-Seq, we cannot overlook a series of possible flaws (to name a few):
- Detection of weakly expressed genes may not be possible,
- RNA-Seq data validation needs to be supported by other methods such as qPCR,
- Needing an available reference genome.
So, which technology should I use for my study?
If the aim is to investigate not only known but also novel transcripts in a gene expression study, then RNA-seq experiments are the most suitable choice. This is why RNA-Seq is ideal for discovery-based experiments; while microarrays are limited to the reference information available during production, RNA-seq experiments may be updated as new sequence information is obtained.
Both the technologies are quite widespread, and there is a large amount of public data available:
The bottom line is, it depends on the answer to these questions:
– Are you interested in lowly expressed genes?
– What kind of data analysis expertise is available?
– How much information is available?
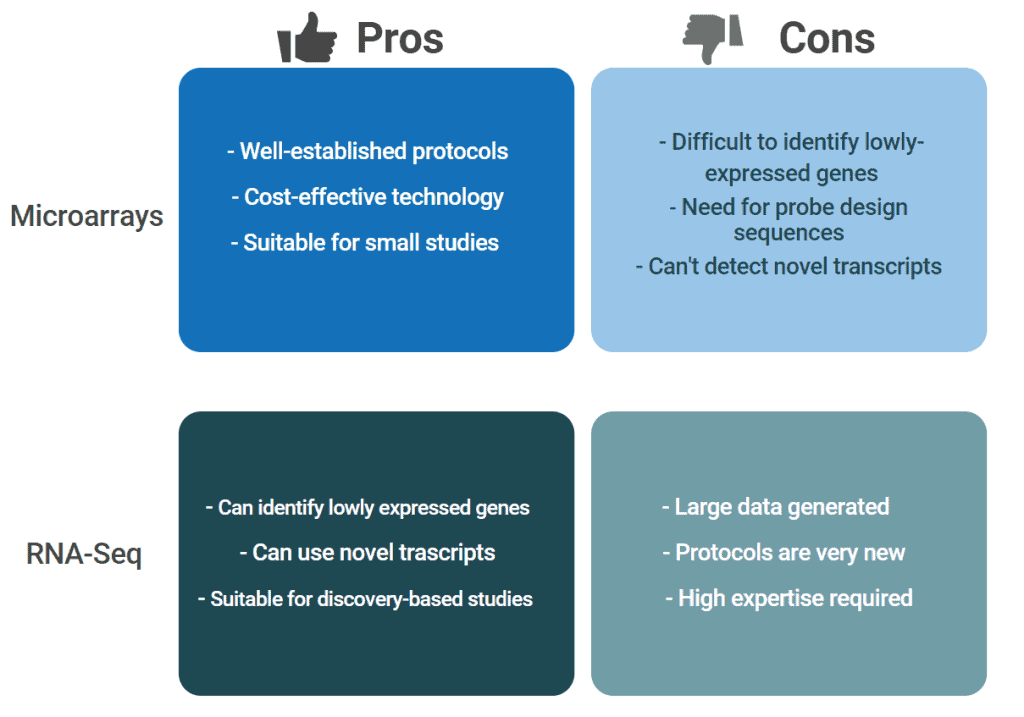
Additionally, pro and cons of the two technologies should be considered:
Publications on RNA-Seq
RNA-Seq is a powerful and versatile tool published widely over the last few years. Below there are highlighted relevant publications to illustrate what work Fios Genomics has been involved in, using both RNA-sequencing and microarrays for different organisations:
- TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells – performed transcriptome RNA sequencing (RNA-Seq) in 298 tissue samples and assessed correlations with PD-L1 expression on immune cells and response.
- Species-specific regulation of angiogenesis by glucocorticoids reveals contrasting effects on inflammatory and angiogenic pathways – identified gene expression changes by next generation sequencing analysis and validated these by real time quantitative PCR of RNA extracted.
- Whole blood gene expression profiling of neonates with confirmed bacterial sepsis – showcased a sequence of analyses carried out on Illumina microarray data.
- Transcriptional changes in chick wing bud polarization induced by retinoic acid – carried out microarray analyses of retinoic acid‐treated wing buds.
Read More
Leave a Reply
You must be logged in to post a comment.

