What is the Future of Microbiome Research?
- 1st August 2019
- Posted by: Claudine Gabriele
- Categories: Bioinformatics, Events, Gene Expression Analysis, Microbiome
After the Microbiome and Probiotics Series: Europe, we caught up with Dr Paul McAdam, one of our Bioinformatics Team Leaders. Watch the video below to find out his thoughts on the microbiome, the future of industry research, and where the big challenges faced will be.
Video Transcript
Q: What is the microbiome and why is research focused on it?
A: The microbiome is basically the population of bacteria that live in a certain niche, whether that is the human gut or on the human skin. There’s growing research in it because there’s increasing evidence that the microbiome plays a role in host health. There are figures that show that we have more bacteria living in our bodies than we have human cells, so it’s not unreasonable to believe that those communities have a role in biology. There’s a growing interest because people have identified an associative role of the microbiome with chronic diseases, such as rheumatoid arthritis, allergies, and asthma. There’s an increasing belief that we can treat the microbiome and have a positive effect on human health.
Q: What does research into the microbiome focus on?
A: So, there’s a lot of focus in microbiome research at the minute on the role of the microbiome in chronic inflammatory disease. There’s evidence to suggest that the microbiome, during infancy, plays a role in asthma and allergies in later life. There’s lots of research going on at the minute to see if we can alter the microbiome, can we treat the microbiome in order to affect the allergy or asthma status of people in later life. There’s also a lot of research going on into the role in neuropsychiatric disorders – again there’s research showing associations of certain compositions of the microbiome with people with schizophrenia and other neuropsychiatric disorders.
Q: What are the biggest research areas for the microbiome?
A: A lot of the recent advances in microbiome research are down to the reduction in the cost of sequencing. Previously, we were only able to sequence a very small, targeted region, the 16S of bacterial strains. Now we use shotgun metagenomic sequencing in order to get a picture of a whole bacterial community within a patient, so we are able to not just look at assigning species, but also look at function, look at what genes are being expressed within the microbiome and that gives us a much better picture of function rather than just composition.
Q: What are the biggest challenges in microbiome research?
A: There’s a number of challenges for microbiome research. If we look at the gut microbiome, there’s a challenge there in actually getting samples. In order to get samples of microbiome from the gut wall, it’s quite an invasive procedure. On top of that, there are many computational challenges. We’re dealing with big data – in many cases, 10s of GBs per sample, and that throws a real computational problem in the way in terms of storage and processing.
Q: Why is sample size for studies so important?
A: Sample size in microbiome studies is just as important as it is for other biological studies, so in order to have the required statistical power to be able to draw definitive conclusions, we need an appropriate sample size so we can be sure that the effects we’re seeing aren’t down to chance. As previously mentioned, there are issues in getting a large sample size in these types of study. Some of these samples can be quite hard to come by, they’re expensive, they’re relatively expensive to generate and the computational challenges for actually processing them once you’ve collected them are quite complex.
Q: Do you think it’s important to share how analysis is done?
A: I think it’s important to share how analysis is done and how the results of research have been achieved. It’s important not only so that scientists can critique people’s methodologies, but also so that people build on those and develop them. Understandably in the commercial sphere, you may not always be able to share your analysis pipelines, that may be the IP that your company’s built on, but in general if analysis pipelines are able to be shared and methodologies are able to be shared, that’s how we ultimately progress in the field.
Q: What are the benefits of microbiome and metabolites parallel analysis?
A: There was a lot of talking about analysing the composition of the microbiome in parallel with the functional profile – so looking at the metabolites or the transcriptomic profile of the metagenome. What that allows, rather than just comparing at the composition or the relative abundance of the bacteria between patients, we can actually look at what metabolites are being produced and how they feed into the host metabolic pathways. That gives us a better handle on the actual underlying biology and how the composition of the microbiome is actually affecting function and how that interplays between the host and the microbiome.
Q: What diseases will benefit from microbiome research?
A: So, there were a lot of presentations on the role of the microbiome in chronic inflammatory conditions, and that seems to be the real area of promise at the minute. There were a lot of presentations looking at the role of the microbiome in asthma, in allergy, in rheumatoid arthritis. A lot of these long-term inflammatory conditions and whether there’s a causative role of the microbiome in that has yet to be elucidated. It could well be that the microbiome changes in response to a disease rather than the other way around, but there’s still further research to be done there.
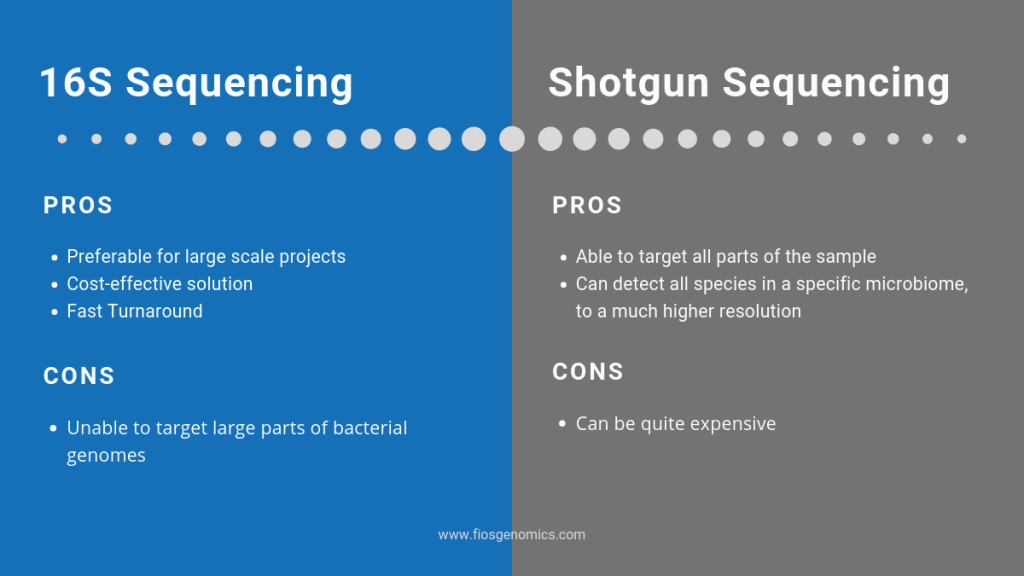
Q: What are the advantages of 16S and shotgun sequencing?
A: So, 16S sequencing is relatively cheap and quick to carry out. The problems are you’re targeting a very small portion of the bacterial genome. You’re not able to amplify 16S from all bacterial species, so there’s by definition going to be strains that you’re missing. As well, bacteria have a very dynamic genome, by just sequencing one small fragment and assigning a species to that, you’re actually missing a large part of information of what the rest of that genome encodes. Shotgun metagenomics gets around this; basically, you sequence everything that’s present in a sample. You get the full picture of what species are actually living in that microbiome and you get that to a much higher resolution, so you’re able to get down to strain level, and you’re able to determine what virulence or resistance genes may be present in that microbiome, and it gives you a much bigger picture to inform our future research.
Q: And what are the disadvantages?
A: The disadvantages to shotgun metagenomics is it can be expensive. It doesn’t have to be if your sample is relatively low diversity and you’ve got low host contamination, but it’s significantly more expensive than 16S in a lot of cases.
Q: What experience does Fios have with microbiome studies?
A: We had a client that was interested in looking at the role of the microbiome during colon cancer progression. They didn’t have the computational experience in house to deal with the data they had generated. We previously successfully completed a large number of projects for them before, so they sent us over the data and we were able to assess it for quality and analyse it in order to determine the microbial composition through different stages of colon cancer, and report any differences back to the client. That informed their future research direction and they decided to take this programme of research further into a larger cohort.
Q: How does data type affect the analysis pipeline?
A: 16S is cheap to generate, it doesn’t generate a large volume of data but downstream we’re not able to analyse the functional aspects of the microbiome in as much detail if we were to use whole metagenome shotgun sequencing. If we use shotgun sequencing, we’re sequencing everything; so much more than we get from 16S, so the file sizes are necessarily much larger, which imposes a larger computational burden, takes longer to analyse, we need more complex pipelines, but also downstream we’re able to get a much better handle on the function of the microbiome. So, we can look at what genes are there rather than just what bacterial species. We can look at what genes are in those strains, we can also do metagenomic transcriptomics or metatranscriptomics, so we can look at the gene expression levels within the microbiome. And then we can also do metabolomic profiling, so we can then look at the metabolites that the microbiome’s producing and link those back to the composition or the gene expression within the microbiome and really get a much better picture of what’s actually going on. So, metagenomic shotgun sequencing gives us the ability to do that. With 16S we’re really limited in terms of the analysis we can do down to the composition of the types of bacteria that are there rather than what function they have.
Q: Where do you see microbiome research going in the future?
A: So, going forward, I think the big advances and challenges in microbiome research are going to be related to whether we can use it as a predictive tool. There’s currently a lot of associations of certain composition of microbiome with diseases such as asthma and type two diabetes. What’s not known is whether the microbiome causes that or whether it’s a side effect of the condition itself, so I think there’s a lot of research effort going in to whether we can use the role of the microbiome to predict these diseases before they happen, so I think a lot of research is going to be focused on that in the medium-term.
Learn More
Interested in finding out how Fios can assist your research? Learn more about the services we offer now.
Leave a Reply
You must be logged in to post a comment.